Analysis enhancements 2024
|
Dear Valued Breeders, Sheep Genetics continually strives to provide a world leading genetic evaluation for our clients. We implement annual analysis enhancements to ensure that we continue to provide world-leading genetic evaluation services to our Sheep Genetics clients. Sheep Genetics works alongside the Animal Genetics and Breeding Unit (AGBU) who are responsible for the cutting-edge research and development behind the enhancements to the Sheep Genetics evaluations. I would personally like to acknowledge the work of the team at AGBU for the many years’ worth of work that have gone into this year’s enhancements. In summary the enhancements implemented this year fall under 2 key themes:
The analysis enhancements will be implemented for the following runs:
This page includes further details about the Analysis Enhancements for 2024. You can also find short videos below with information about this year’s enhancements. Please review these if you have any questions, alternatively you can contact one of the Sheep Genetics team. Your Sincerely, Peta Bradley |
Analysis enhancement videos
Summary of the upcoming enhancements for each relevant analysis.
| Analysis enhancement | MERINO | TERMINAL | MATERNAL | What this means for you |
|
Inclusion of objective carcase data collected from processors |
X |
X |
X |
Sheep Genetics will be able to use data from collected in plants from DEXA and MEQ/SOMA technologies to inform carcase ASBVs (LMY, & IMF). |
X |
X |
X |
Updates to data quality control steps for the shear force at 5 days analysis will be applied, removing data where a contemporary group has an average shear force >50nM. | |
|
Transforming carcase data to standard contemporary group mean |
X |
X |
X |
Carcase data will be transformed to a standard contemporary group mean for DRESS, LMY, IMF, and SHEARF5 to account for differences in variation across slaughter groups. |
|
Genomic breed reference update (includes new breeds and allele frequencies) |
X |
X |
The genomic reference has been expanded to allow more breeds to benefit from the genomics, resulting in more accurate ASBVs. | |
|
3. |
X |
The MERINOSELECT research indexes which were released in May 2023 have been adjusted following industry consultation and will be finalised as commercial selection indexes. |
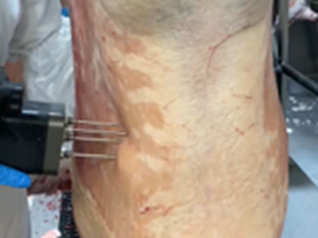
Enhancement 1.1 - Inclusion of objective carcase data collected from processors
In conjunction with processors across Australia adopting objective measurement technology in plant, including additions of dual x-ray absorptiometry (DEXA) machines to measure lean meat yield (LMY), as well as meat eating quality (MEQ) probes and SOMA devices to measure intramuscular fat (IMF). Sheep Genetics is now in position to work with breeders to utilise objective carcase data collected from processors which have accredited carcase and eating quality devices in plant to inform LMY and IMF ASBVs in the genetic evaluation.
Currently, there are over 1,500 phenotypes for IMF and almost 5,500 phenotypes for LMY collected in plant which will now be able to inform carcase and eating quality breeding values. Previous research from the ALMTech projects showed that the genetic correlations between carcase data collected in plant and the gold standard measures of carcase data (CT machine for LMY, and chemical IMF) are high to very high. This ultimately means that data collected from these objective technologies can be used within the Sheep Genetics evaluations to inform carcase and eating quality ASBVs.
Allowing data collected in plant from these objectives technologies provides another pathway for breeders to submit carcase data to inform ASBVs on their animals. However, it’s important to note that measures for shear force are currently unable to be collected in plant, so it’s still important to phenotype for shear force (via sire nominations to the resource flock). Also, as hook tracking accuracy issues still persist in plant, Sheep Genetics and AGBU are applying data quality control steps to the data prior to submission into the genetic analysis to ensure that the data entering the analysis is accurate.
 |
 |
| Figure 1. MEQ | Figure 2. SOMA |
If you have a consignment of animals that meets these requirements (see below) and are interested in being involved, please contact Sheep Genetics before the animals are consigned to ensure that all the necessary information is collected. Please also contact the processor to inform them that the consignment will involve the collection of data for the genetic analysis and to keep an eye on hook tracking. The below are requirements for breeders who are wanting to collect commercial carcase measurements:
- Animals must have a Sheep Genetics ID and are in the Sheep Genetics evaluation (either excess animals from the stud or have pedigree information)
- A pre-slaughter weight (4 hours fasted) on-farm prior to slaughter. Ideally pre-slaughter weight is to be recorded as close as possible to the animals going onto the truck to slaughter, however, pre-slaughter measurements can be taken up to 7 days prior to slaughter
- Please supply, date of pre-slaughter measurements, on-farm management group of animals, kill date, and consignment number
- Where possible, we recommend taking the following on farm pre-slaughter measurements also
- Condition score
- Fat and muscle scan data
Please contact Sheep Genetics for a template for the pre-slaughter data. Following the kill, please provide Sheep Genetics with the unedited kill sheet direct from the processor, as well as the required pre-slaughter data in the appropriate template.
Meat science data will still be recorded on the resource flock to ensure representation of industry sires are phenotyped and recorded for all meat science traits, particularly Shear Force at 5 days which currently cannot be measured in plant.
|
Who is impacted? Both LAMBPLAN and MERINOSELECT breeders will be able to submit data collected from processors to the genetic analysis. The addition of more phenotypes for IMF and LMY will result in movement in ASBVs across both LAMBPLAN and MERINOSELECT analysis for carcase and eating quality ASBVs and correlated traits. Please contact Sheep Genetics prior to the consignment to ensure all pre-slaughter requirements are taken. |
Enhancement 1.2 - Shear force validation
Shear force at 5 days (measured in Newton metres, nM) measures the energy or force required to cut through a loin muscle after aging for 5 days. Shear force measures meat toughness, and is therefore, an indicator of meat eating quality. Shear force is measured in the loin of lambs, with the majority of data recorded on lambs killed through the Resource Flock or specific eating quality progeny tests, which is then used to inform ASBVs for SHEARF5.
The Animal Genetics and Breeding Unit (AGBU) have been exploring the shear force analysis to improve how the analysis is accounting for the impact of non-genetic factors, such as cold shortening. Removing contemporary groups with an average shear force value above 50Nm was shown to lead to improvements in heritability and validation metrics. This indicates that for contemporary groups with a shear force average above 50nM the phenotypes collected are not a reflection of the animals genetic merit and should be excluded from the evaluation.
The data quality control steps for SHEARF5 have been updated to now exclude phenotypes from the contemporary groups with an average shear force greater than 50 nM. This exclusion means there will be better filtering of outliers for shear force phenotypes, as well as an improved heritability for SHEARF5 as phenotypes with significant non-genetic impacts on performance are being filtered out.
|
Who is impacted? There will be movement in SHEARF5 ASBVs for animals in LAMBPLAN and MERINOSELECT. Terminal selection indexes (LEQ, EQ, and TCP) will also be influenced from the updated quality control steps for shear force data. |
Enhancement 1.3 – Transforming carcase data to a standard contemporary group mean
Data for all carcase traits, DRESS, LMY, IMF, and SHEARF5 will now be transformed to a standard contemporary group mean. This update is being applied to account for differences in variation in trait expression across kill groups. This technique is currently used for many traits within the evaluation with the changes from this update beneficial but minimal in the observed outcome. As the analysis of the carcase ASBVs will be updated, there may be movement in animal’s ASBVs for DRESS, LMY, IMF, and SHEARF5 in LAMBPLAN and MERINOSELECT as well as Terminal (LEQ, EQ, and TCP) and Merino (ML and SM) selection indexes.
|
Who is impacted? This enhancement will be applied to both LAMBPLAN and MERINOSELECT analysis, influencing Terminal, Maternal, and Merino animals. Individual animals ASBVs for carcase traits like DRESS, LMY, IMF, and SHEARF5 in the Terminal, Maternal, and Merino analyses will be influenced, as well as Terminal selection indexes (LEQ, EQ, and TCP). |
Enhancement 2 - Genomic breed reference
Genomics uses the mapping of DNA to accurately identify common genes, and therefore is able to more accurately identify relationships between animals in Sheep Genetics. This allows for more accurate ASBVs using a single-step, particularly where a genotype is taken early in life and well linked to a reference population, for traits that are hard to measure, measured later in life or on carcases, or are sex limited.

Figure 3. Genomics is used to accurately identify relationships between animals, hence increasing the accuracy of ASBVs.
The genomic pipeline was first introduced into Sheep Genetics in 2017, and since then, both breeds and traits with access to single-step breeding values have been updated over time. The massive uptake of genotyping across many breeds/populations has resulted in a rapid growth in the genomic reference populations (groups of animals with both a genotype and a range of phenotypes). With the expansion of the genomic reference, we are now able to include a large number of genotypes from new breeds/populations and composites within LAMBPLAN with the 2024 analysis enhancements. This update is not occurring in MERINOSELECT or DOHNES.
Previously, the adequacy of reference populations was determined at a breed level, with 13 breeds receiving genomically enhanced ASBVs in LAMBPLAN. Reference populations are now identified at a population level, meaning that more composite breeds and new breeds will have access to single step ASBVs.
Over 29,000 and 13,000 new genotypes will be included in the LAMBPLAN Terminal and Maternal analyses from the 15th of May run. These new genotypes are associated with animals from breeds/populations that previously did not have access to genomically enhanced ASBVs. The inclusion of new genomic information may have a relatively large impact on ASBVs and selection indexes across the LAMBPLAN analyses. It will have the most impact on animals and flocks that have animals which are now having their genotype included in the calculation of ASBVs. It will also have an impact on animals that are closely related to the animals gaining a genotype, primarily in currently genotyped individuals. The changes in ASBVs are associated with an increase in the information contributing to the estimation of the ASBV.
The inclusion of genomic information into breeding values will result in movement in ASBVs and selection indexes, and some re-ranking of animals. The addition of a genotype into the analysis for an animal does not result in better ASBVs, only more accurate ASBVs. Therefore, this enhancement is equally likely to be favourable as unfavourable, with genomic information increasing the accuracy of relationships between animals, making the breeding values more accurate, not necessarily more favourable.
Following the 15th of May analysis run in LAMBPLAN, the genotypes included in the analysis reflect the diversity of breeds within the Sheep Genetics sheep population. The ability to use genomic information is still reliant on an adequate genomic reference for your animals and flock. This reference needs to be maintained by recording relevant phenotypes on genotypes animals. Examples of populations that will now receive genomically enhanced breeding values include Ultra White, Australian Whites, Composite Shedders, a range of Composite Terminals and more. If you are unsure if your sheep will have an adequate reference population for traits in your breeding objective, please contact Sheep Genetics.
|
Who is impacted? All Maternal and Terminal Breeders Particularly those who will have new genotypes included with analysis enhancements 2024. |
Enhancement 3 - MERINOSELECT Indexes
Indexes are a useful tool when making selection decisions. They are best used when initially ranking animals for several traits quickly and easily, to remove the lower portion of animals, and rank based on individual traits of importance to your breeding objective for final selection decisions. Indexes are designed to meet the diversity of industry breeding objectives and production systems, both for commercial and seedstock producers.
In June of 2023 Sheep Genetics released 5 new MERINOSELECT research indexes. This was followed by a period of collecting and reviewing industry feedback on the new indexes to help inform the transition pathway from research indexes to industry indexes for the MERINOSELECT analysis.
Based on the feedback there were changes made, resulting in some differences between the research and finalised selection indexes. The changes that occurred from research to final version for 2024 will be discussed below. Documentation further outlining the indexes and their production systems will become available on the Sheep Genetics website to assist in selecting an index best suited to your breeding objective and production system.
From the 21st of May MERINOSELECT run, there will be 4 standard selection indexes for Merinos; Fine Wool (FW), Wool Production (WP), Sustainable Merino (SM) and Merino Lamb (ML). Below is a summary of the pathway we have taken from the old to research to final indexes.
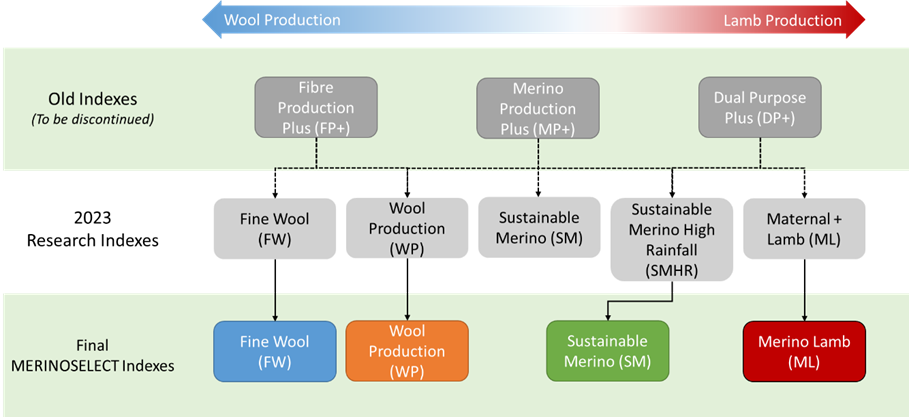
Figure 3. Pathway of Index development from June 2023 to May 2024
Table 2 below outlines the changes from researched to final versions is included below. More information on the changes for each index will be found in documentation on the Sheep Genetics website.
Table 2. Changes between the research and final indexes.
| Final Index Name | Research Index Name | Summary of index | Changes from Research to Final versions |
| Fine Wool (FW) | Fine Wool |
Production system where the majority of income is from wool clip, with a strong focus on the quality of the clip. Producing 15-17 micron wool from both the breeding flock and a mixed age wether flock. The wool to meat income ratio of the modelled production system is 75:25. Genetic improvement of fleece weight, fibre diameter, staple strength and reproduction. Desired gain approach to provide weighting on reducing wrinkle and worm egg count. |
|
| Wool Production (WP) | Wool Production |
Production system where the majority of income is from wool clip, with a strong focus on the quality of the clip. Producing 16-18 micron wool from both the breeding flock and a mixed age wether flock. The wool to meat income ratio of the modelled production system is 71:29. Genetic improvement of fleece weight, fibre diameter, staple strength and reproduction. Desired gain approach to provide weighting on reducing wrinkle. |
|
| Sustainable Merino (SM) | Sustainable Merino High Rainfall |
Production system where the income is from sheep meat production and the wool clip are reasonably balanced, but there still remains a large focus on improving the wool clip (quantity and quality). Producing 17-19 micron wool and selling lambs at a post-weaning age stage, off shears. The wool to meat income ratio of the modelled production system is 46:54. Genetic improvement of fleece weight, growth and lean meat yield and reproduction. A desired gain approach to provide weighting on reducing wrinkle, dag and worm egg count. |
|
| Merino Lamb (ML) | Maternal Lamb |
Production system where the majority of income comes from sheep meat, primarily from fast growing lambs, but the wool clip from adult ewes still remains an important part of the production system (18-21 micron). The wool to meat income ratio of the modelled production system is 31:69. Genetic improvement of fleece weight, growth, lean meat yield, eating quality and reproduction. A desired gain approach to provide weighting on reducing wrinkle. |
|
|
Who is impacted? Merino breeders using indexes for selection decisions. Ensure to look at the relevant information for which of the final indexes is the best suited for your breeding objective. |